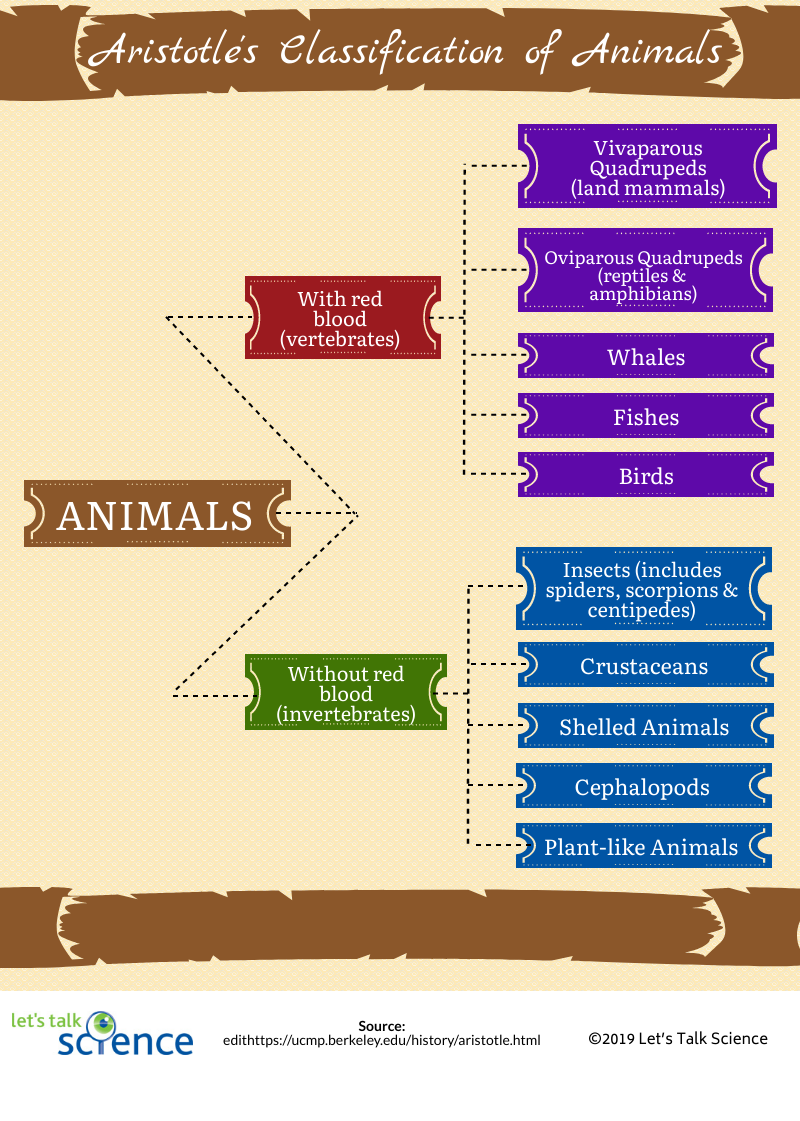
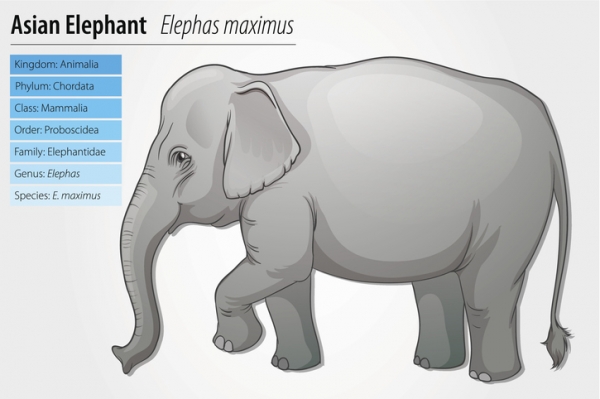
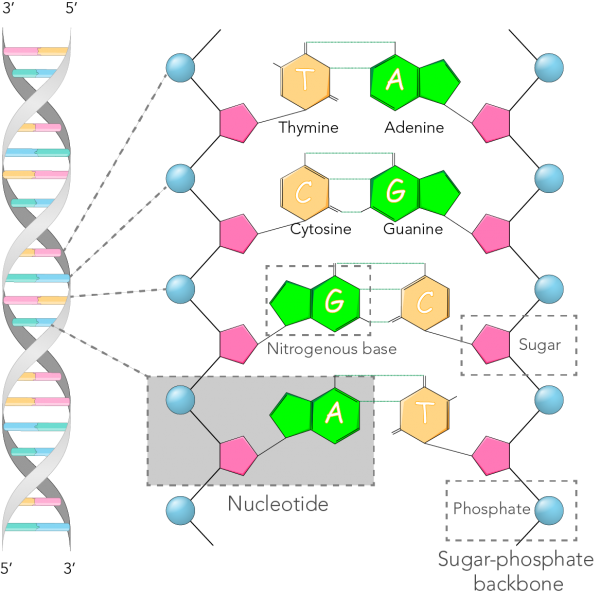
The Classification of Life: From Linnaeus to DNA Barcoding
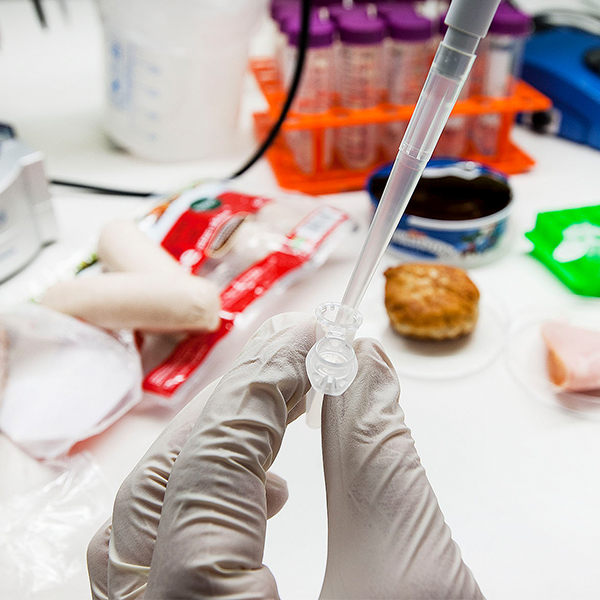
DNA barcoding used to test food (NTNU Vitenskapsmuseet [CC BY], Wikimedia Commons)

DNA barcoding used to test food (NTNU Vitenskapsmuseet [CC BY], Wikimedia Commons)
8.1
How does this align with my curriculum?
AB
10
Knowledge and Employability Science 10-4 (2006)
Unit D: Investigating Matter and Energy in Environmental Systems
AB
10
Science 14 (2003, Updated 2014)
Unit D: Investigating Matter and Energy in Environmental Systems
BC
11
Environmental Science 11 (June 2018)
Big Idea: Complex roles and relationships contribute to diversity of ecosystems.
NU
10
Knowledge and Employability Science 10-4 (2006)
Unit D: Investigating Matter and Energy in Environmental Systems
NU
10
Science 14 (2003, Updated 2014)
Unit D: Investigating Matter and Energy in Environmental Systems
YT
11
Environmental Science 11 (British Columbia, June 2018)
Big Idea: Complex roles and relationships contribute to diversity of ecosystems.
NT
10
Knowledge and Employability Science 10-4 (Alberta, 2006)
Unit D: Investigating Matter and Energy in Environmental Systems
NT
10
Science 14 (Alberta, 2003, Updated 2014)
Unit D: Investigating Matter and Energy in Environmental Systems